Abstract:
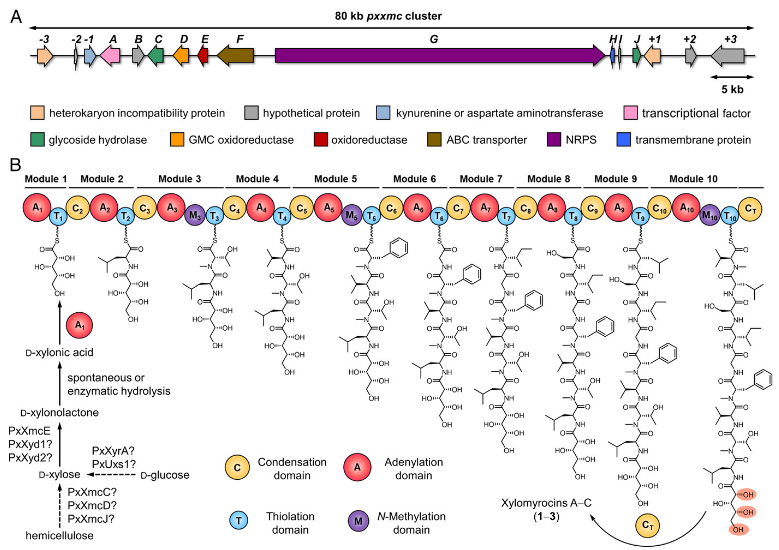
Xylomyrocins, a unique group of nonribosomal peptide secondary metabolites, werediscovered inParamyrotheciumandColletotrichumspp. fungi by employing a combina-tion of high-resolution tandem mass spectrometry (HRMS/MS)–based chemometrics,comparativegenomemining,genedisruption,stableisotopefeeding,andchemicalcomplementation techniques. These polyol cyclodepsipeptides all feature an unprece-dentedD-xylonic acid moiety as part of their macrocyclic scaffold. This biosynthon isderived fromD-xylose supplied by xylooligosaccharide catabolic enzymes encoded in thexylomyrocinbiosyntheticgenecluster,revealinganovellinkbetweencarbohydratecatabolism and nonribosomal peptide biosynthesis. Xylomyrocins from different fungalisolates differ in the number and nature of their amino acid building blocks that areneverthelessincorporatedbyorthologousnonribosomalpeptidesynthetase(NRPS)enzymes. Another source of structural diversity is the variable choice of the nucleophilefor intramolecular macrocyclic ester formation during xylomyrocin chain termination.This nucleophile is selected from the multiple available alcohol functionalities of thepolyol moiety, revealing a surprising polyspecificity for the NRPS terminal condensa-tion domain. Some xylomyrocin congeners also featureN-methylated amino acid resi-duesinpositionswherethecorrespondingNRPSmodules lackN-methyltransferase(M) domains, providing a rare example of promiscuous methylation in the context ofan NRPS with an otherwise canonical, collinear biosynthetic program.
Keywords: