Recently, the latest research of the microbial functional genomics team led by Professor Min Lin of Biotechnology Research Institute, CAAS indicated that a novel regulatory noncoding RNA is cooperatively involved in the optimal expression of nitrogenase genes in Pseudomonas stutzeri A1501. It lays an important theoretical foundation for further revealing the regulation mechanism of biological nitrogen fixation network. The related result was online published in the latest issue of the Applied and Environmental Microbiology.
Biological nitrogen fixation is a unique physiological function of nitrogen-fixing bacteria, which is catalyzed by nitrogenase, and is greatly influenced by intracellular energy supply and environmental stress factors. To adapt to diverse environments, nitrogen-fixing bacteria have evolved various regulatory mechanisms to control the transcription of nif genes and maintain the efficient nitrogenase activity, yet the underlying mechanism is not fully understood.
The researchers have previously shown that a novel regulatory noncoding RNA NfiS in Pseudomonas stutzeri A1501. This study reported the identification and characterization of a second ncRNA involved in the post-transcriptional regulation, termed NfiR. Phenotypic analysis and proteomic analysis of nfiR mutant showed that NfiR also played an important regulatory role in environmental stress response and nitrogen fixation, and many potential regulatory targets were identified. Results of microscale thermophoresis and genetic complementation strongly suggest that nifD mRNA is a direct target of NfiR. Phenotypes of the double deletion mutant lacking nfiR and nfiS suggest that NfiR, in concert with NfiS, optimizes nitrogenase production at the posttranscriptional level. This work demonstrated for the first time that two non-coding RNAs responding differently to various environmental signals coordinate the regulation of nitrogenase activity, which may be an evolutionary adaptation strategy of nitrogen-fixing bacteria in complex and changeable rhizosphere environment.
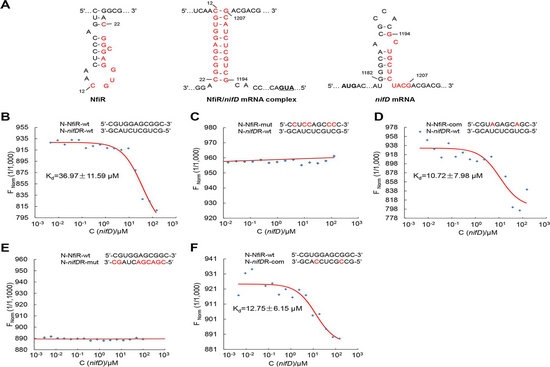
Molecular interactions between NfiR and nifD mRNA
Compared with Rhizobium nitrogen fixing system, the rice root-associated bacteria do not form nodules and other special organizational structure, and was greatly affected by the environment. Its field nitrogen fixation efficiency was low and its application effect was unstable. To overcome the above natural shortcomings, it is a challenging research topic to establish an efficient and stable associative nitrogen fixation system and its application in agricultural production. This study has taken a solid step to solve this problem.
This work was supported by the National Basic Research Program of China and National Science Foundation of China. Dr. Yuhua Zhan and Zhiping Deng are the co-first authors of the paper. Professor Qi Cheng and Min Lin are the co-corresponding author.
Link: https://aem.asm.org/content/85/14/e00762-19
|