Recently, Prof. Xiaofeng Gu’s lab from the Biotechnology Research Institute, Chinese Academy of Agricultural Sciences (CAAS), and his collaborators have profiled the single-cell-resolution transcriptome landscape of root tissue in two monocot model plant rice cultivars (japonica ‘Nipponbare’ and indica ‘93-11’), opening new avenues to study the specialization, function and evolution of plant cell types. The results have been published on Molecular Plant.
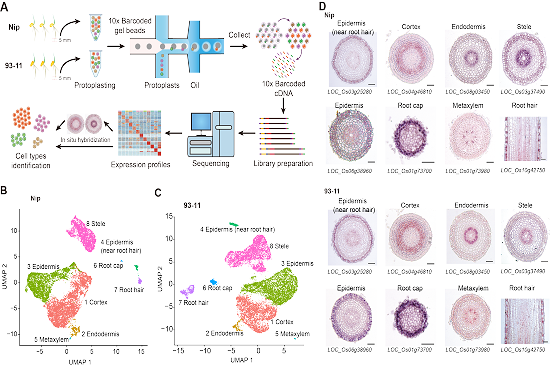
Throughout the life cycle of plants, the root tips are constantly differentiating and developing. The formed roots absorb water and mineral elements from the soil, participate in the interaction between plants and biological or abiotic signals, and contribute to fixing plants. However, little is known about these programs at single cell resolution in plants. We performed single-cell RNA sequencing (scRNA-seq) of 10,968 and 12,564 cells isolated from Nip and 93-11root tips, and identified 8 unique cell types. Functional enrichment analysis was performed on the differentially expressed genes of each cell type, and our data suggest that most genes are related to stimuli responses, indicating that Nip and 93-11 have different response mechanisms to external stimuli.
Through the pseudo-time analysis of the single cell data, the development trajectory of root epidermal cells was plotted. Cells from the two cultivars showed highly consistent pseudotime order, suggesting a conserved developmental trajectory. We further combined rice and Arabidopsis datasets and aligned them by using orthologous genes, which was the first time to analyze the evolution of plant tissues and organs at the single cell level. Great difference was found between these two species, whereas cell type of root hair is conserved between rice and Arabidopsis. The results provide insight into the genetic basis of root type differentiation in monocotyledons and dicotyledons at single-cell resolution.
Prof. Xiaofeng Gu and Jan U. Lohmann (Heidelberg University) are the corresponding authors. This work was supported by National Transgenic Major Program (2019ZX08010-002) (to X.G.), Central Public-interest Scientific Institution Basal Research Fund (No. Y2020PT06) (to X.G.), and the intramural research support from Chinese Academy of Agricultural Sciences.
Original link:
https://www.sciencedirect.com/science/article/pii/S1674205220304457
|