A research team from the Chinese Academy of Agricultural Sciences (CAAS) has developed a novel strategy for predicting and designing high-level protein expression, powered by a pre-trained protein language model. The study, a collaboration between the Agricultural Microbial Protein Design and Intelligent Manufacturing Innovation Team from the Institute of Biotechnology and the Microbial and Enzyme Engineering Innovation Team from the Institute of Animal Science, was recently published in Advanced Science.
Efficient soluble heterologous expression is crucial for translating enzymatic proteins into commercial products. Conventional strategies, such as switching expression hosts, vectors, or employing molecular chaperones, often rely heavily on researcher experience and require extensive experimental validation. To address this, the team developed MP-TRANS, a domestically built pre-trained protein language model, leveraging transfer learning theory. They innovatively introduced the concepts of the Amino acid Expression Index (AEI) and the Strength of Relative Amino acid Bias (SRAB), providing precise quantitative tools for analyzing protein expression.
By fine-tuning the MP-TRANS model for specific downstream tasks, the researchers constructed two specialized models: an expression level predictor (MPB-EXP) and a mutant generator (MPB-MUT). Notably, MPB-EXP stands as the current prediction model supporting the largest number of expression hosts (88 species), achieving an average prediction accuracy of 0.78. Experimental validation confirmed that the soluble expression levels of xylanase, cellulase, and PET-degrading enzymes in E. coli were significantly enhanced using this strategy. This work, which deeply integrates large protein language models with genetic expression principles, offers a new paradigm and powerful tool for the efficient creation of high-performance protein products.
This research was supported by the National Key R&D Program of China, the National Natural Science Foundation of China, and the CAAS Agricultural Science and Technology Innovation Program. Computational resources were provided by the Hebei Artificial Intelligence Computing Center.
The corresponding authors of the paper are Dr. Jian Tian and Dr. Huoqing Huang from the Institute of Animal Science, CAAS, and Dr. Feifei Guan and Dr. Bo Liu from the Institute of Biotechnology, CAAS. Graduate students Tuoyu Liu and Yiyang Zhang are the co-first authors.
Full article URL:
https://onlinelibrary.wiley.com/doi/10.1002/advs.202407664
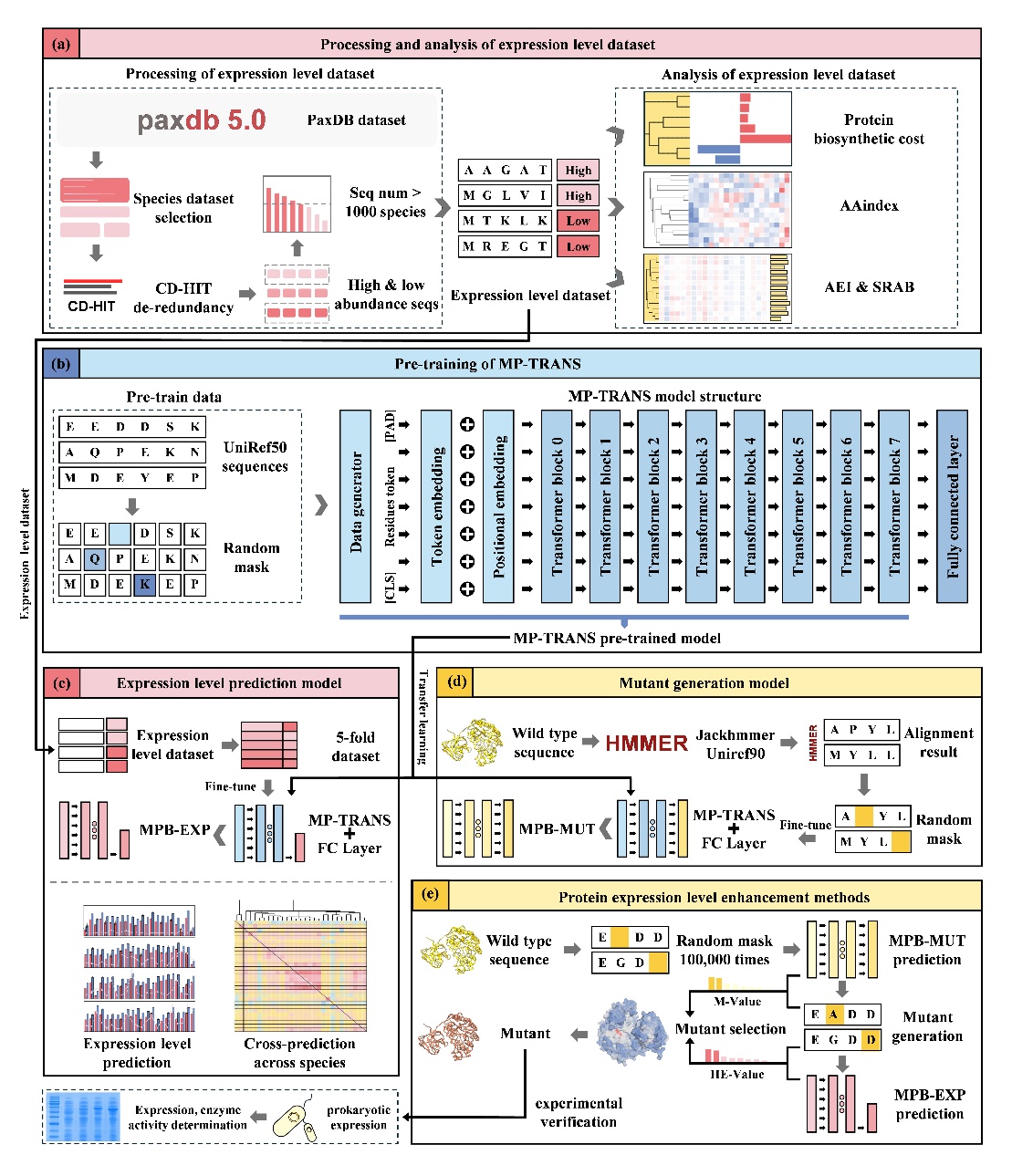
Workflow to predict and generate mutants to enhance soluble expression of proteins |