Recently, Protoplast transient expression-based RNA-sequencing (PER-seq): A simple method to screen transcriptional regulation in plants was developed by the Crop Metabolic Regulation and Nutrient Enhancement Innovation team of the Biotechnology Research Institute. PER-seqdoes not require mutants or transgenic lines, provides a novel and efficient tool for transcriptional regulatory network analysis, mining of transcription factor target genes, and predicting gene regulatory pathways. The relevant results are published in “Plant Physiology”.
Screening the target genes of transcription factors plays a key role in elucidating the mechanisms of plant development and stress resistance regulation. Conventional RNA-sequencing (RNA-seq) and chromatin immunoprecipitation sequencing (ChIP-seq) are limited by the availability of genetic materials. DNA affinity purification sequencing (DAP-seq) and cleavage under targets and tagmentation (CUT&Tag) technologies can identify transcription factor binding sites, but cannot determine transcriptional activation or inhibition. At present, there is still a lack of simple, rapid and widely applicable target gene screening methods for transcription factors.
Opaque2 (O2) is a basic leucine zipper family TF crucial for maize (Zea mays) grain filling. Moreover, recent studies reported that O2 participated in seed size regulation through the O2-ZmGRAS11 module (JIPB 2021). However, ZmGRAS11 was not identified as an O2 target through previous genome-wide screening, indicating that the regulatory function of O2 remains to be fully explored. A comprehensive understanding of the regulatory role of O2 is of great significance for achieving synergistic and precise regulation of grain quality and yield traits.
In response to the above challenges and problems, PER-seq technology was developed.Moreover, the target genes of O2 were screened and several new target genes were found, which revealed and expanded the potential functions of O2 in synergistic storage substance synthesis, sugar and micronutrient transport, grain development and stress response. The application of PER-seq in plants such as wheat and Arabidopsis thaliana has proved that it is widely adaptable and fast, independent of mutant or transgenic plants. In addition, PER-seq technology is particularly suitable for constructing transcriptional regulatory networks for key transcription factors that cause mutation death.
Guest master JiamengZhu (currently a doctoral student at Nanjing Normal University) and Dr. SuzhenLi are co-first authors of this research.ProfessorXiaojinZhou is the corresponding author. Rumei Chen and Baobao Wang (Biotechnology Research Institute) and Haiyang Jiang (Anhui Agricultural University) participated in this work. This study was supported by grants from the National Key Research and Development Program of China (2021YFF1000300), the National Natural Science Foundation of China (32272118), and the National Special Program for GMO Development of China (2016ZX08003-002).
More information can be found through the link:
https://doi.org/10.1093/plphys/kiad495
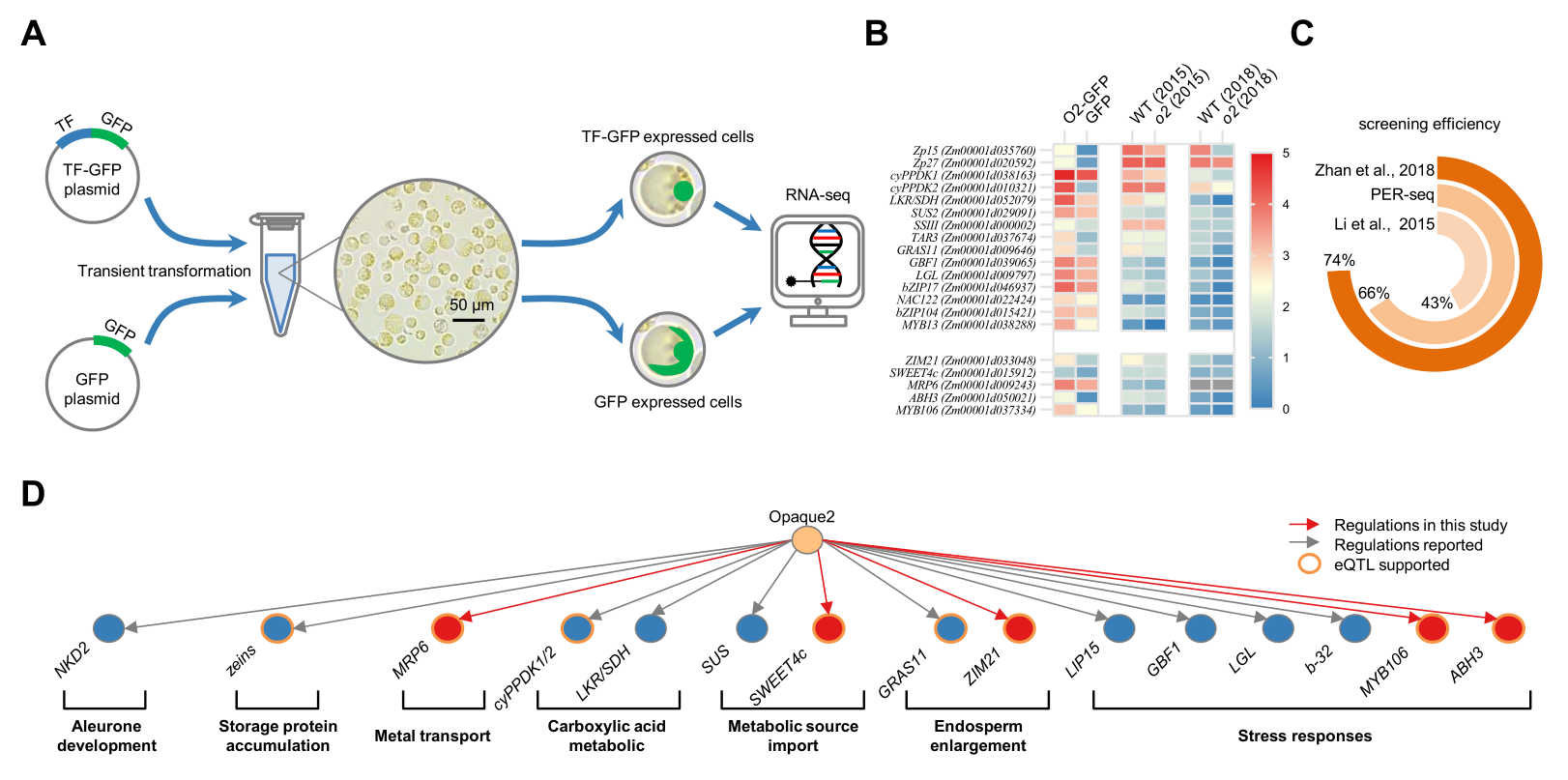
Fig 1. Identification of O2 targets using the PER-seq technique
The red arrows represent the newly obtained target genes, the gray arrows represent the known target genes, and the orange borders represent the transcriptional regulatory effects supported by eQTL data. |