Recently, Gu Xiaofeng's lab, Tian Jian's lab, and Pu Li's lab from the Institute of Biotechnology, Chinese Academy of Agricultural Sciences published a titled " A deep learning approach to automate whole-genome prediction of diverse epigenomic modifications in plants" research paper, constructed SMEP (http://www.elabcaas.cn/smep/index.html), an online tool for intelligent prediction of plant epigenomic modifications. This work uses artificial intelligence (AI) methods to deeply learn plant DNA methylation, RNA methylation, histone modification and other sequence information, and systematically realize the apparent modification sites in rice, maize and other species Prediction, providing tools and data support for crop functional genomics research and intelligent design and breeding.
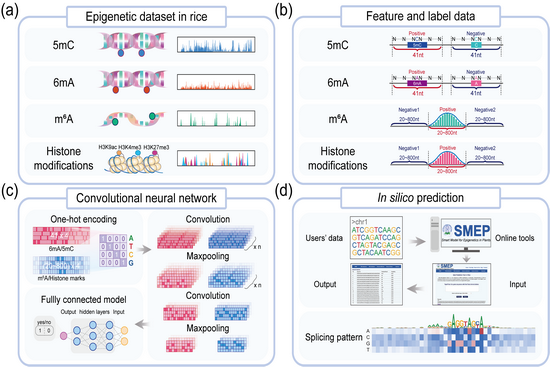
Epigenetic modifications include DNA/RNA methylation and histone modification, which are the main driving factors regulating eukaryotic gene transcription, RNA metabolism and other biological processes. In recent years, the development of detection technology and high-throughput sequencing has promoted the research of crop epiomics. However, due to the detection technology, experimental cost, material organization and dynamic and reversible regulation characteristics of apparent modification, there are still a large number of apparent modification sites that have not been explored and studied. Therefore, how to use the existing epiomics data to in-depth explore potential epigenetic modification sites is a key issue facing current epigenetic research.
At present, artificial intelligence technology has shown broad application potential in epiomics big data analysis and prediction. The research is based on the convolutional neural network (CNN) method and used the early stage of the research team to draw a variety of apparent modification maps of japonica rice Nipponbare, and constructed a highly accurate intelligent prediction model (SMEP). After parameter optimization, cross-validation and experimental verification, SMEP has high confidence in predicting epigenetic modification sites such as DNA methylation, RNA methylation and histone modification. At the same time, the study also successfully applied the SMEP model to the prediction of apparent modification sites in the representative indica rice variety 93-11 and the maize line B73. Since SMEP predicted a large number of sites that could not be detected under normal experimental conditions, the team further verified the 6mA modified sites predicted by SMEP to specifically respond to heat shock through heat shock stress treatment experiments, indicating the apparent modified sites predicted by SMEP Points may participate in multiple environmental responses. Finally, an SMEP intelligent prediction tool was built online, providing a visual interface for searching epigenetic modification sites and gene expression data for user reference, and providing mining tools and data support for the intelligent design of important agronomic traits of rice, maize and other species.
The Institute of Biology Gu Xiaofeng, Tian Jian, and Puli are the corresponding authors of the paper, and graduate students Wang Yifan, Zhang Pingxian, Guo Weijun, and Liu Hanqing are the co-first authors. The research has won the National Genetically Modified Major Project, the National Natural Science Foundation of China, the Science and Technology Innovation Project of the Chinese Academy of Agricultural Sciences, etc. Project funding.
|