Recently, Dr. Zhe Liang’s group from Biotechnology Research Institute of Chinese Academy of Agricultural Sciences,present a comprehensive quantitative analysis of the proteome across 14 major rice tissues, revealing that N6-methyladenosine (m6A) is negatively correlated with protein abundance. This finding provides insight into the longstanding discrepancy observed between RNA and protein levels in plants. The work was published as a Research Article in Nature Plants.
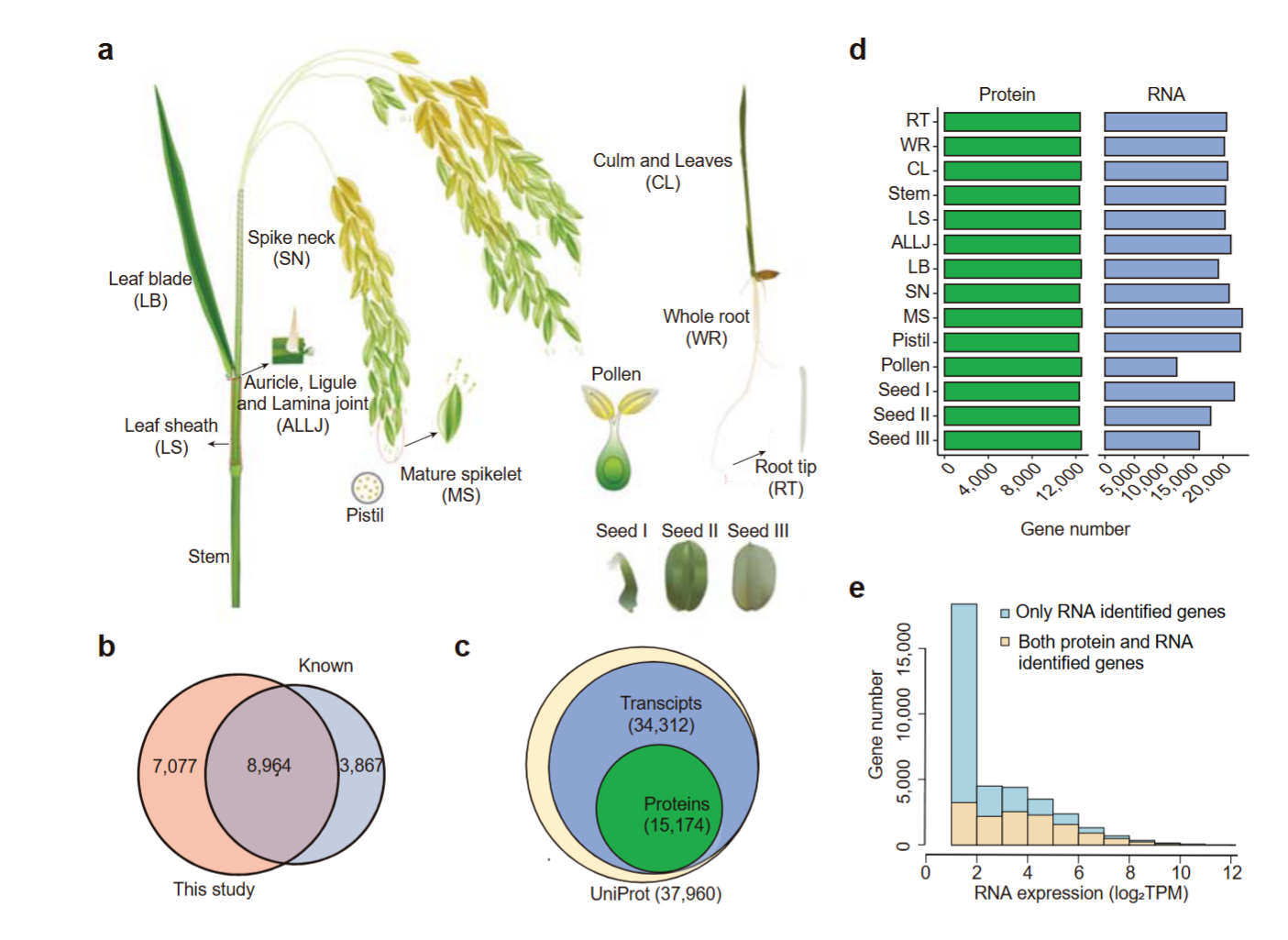
Rice is one of the most important staple food and model species in plant biology, yet its quantitative proteomes are largely uncharacterized. This study quantifies the relative protein levels of over 15,000 genes across major rice tissues using atandem mass tag strategy followed by intensive fractionation and mass spectrometry.This study identifiestissue-specific and -enrichedproteins that are linked to the functional specificity of individual tissues.Proteogenomic comparison of rice and Arabidopsis reveals conserved proteome expression, which differs from mammals in that there is a strong separation of speciesrather than tissues. Notably, profiling of N6-methyladenosine (m6A) across the rice major tissues shows that m6A at untranslated regions is negatively correlated with protein abundance and contributes to the discordance between RNA and protein levels. This work also demonstrate that our data are valuable for identifying novel genes required for regulating m6A methylation. Taken together, this study provides a paradigm for further research into rice proteogenome.
This work was supported byBiological Breeding-Major Projects (2023ZD04076, 2022ZD0401904), National Natural Science Foundation of China (3217040580), The Agricultural Science and Technology Innovation Program (ASTIP), Central Public-interest Scientific Institution Basal Research Fund,and Central Laboratory of Biotechnology Research Institute, CAAS.
|